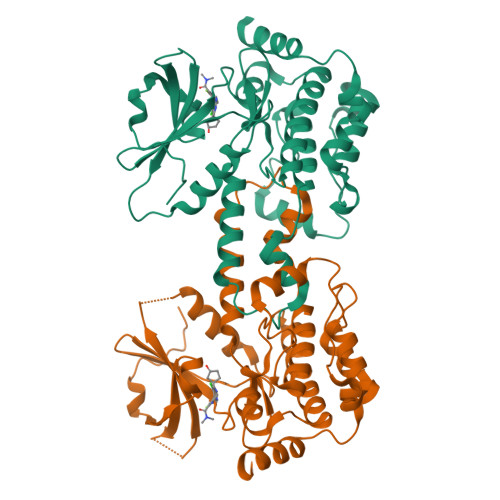
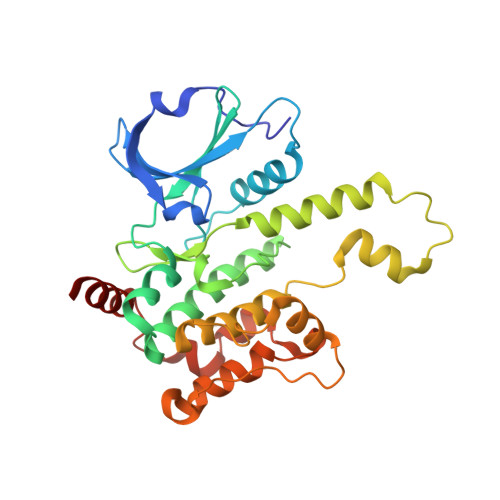
Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Chan, B.K., Seward, E., Lainchbury, M., Brewer, T.F., An, L., Blench, T., Cartwright, M.W., Chan, G.K.Y., Choo, E.F., Drummond, J., Elliott, R.L., Gancia, E., Gazzard, L., Hu, B., Jones, G.E., Luo, X., Madin, A., Malhotra, S., Moffat, J.G., Pang, J., Salphati, L., Sneeringer, C.J., Stivala, C.E., Wei, B., Wang, W., Wu, P., Heffron, T.P.(2022) ACS Med Chem Lett 13: 84-91
- PubMed: 35059127
- DOI: https://doi.org/10.1021/acsmedchemlett.1c00473
- Primary Citation of Related Structures:
7R9L, 7R9N, 7R9P, 7R9T - PubMed Abstract:
Hematopoietic progenitor kinase 1 (HPK1) is implicated as a negative regulator of T-cell receptor-induced T-cell activation. Studies using HPK1 kinase-dead knock-in animals have demonstrated the loss of HPK1 kinase activity resulted in an increase in T-cell function and tumor growth inhibition in glioma models. Herein, we describe the discovery of a series of small molecule inhibitors of HPK1. Using a structure-based drug design approach, the kinase selectivity of the molecules was significantly improved by inducing and stabilizing an unusual P-loop folded binding mode. The metabolic liabilities of the initial 7-azaindole high-throughput screening hit were mitigated by addressing a key metabolic soft spot along with physicochemical property-based optimization. The resulting spiro-azaindoline HPK1 inhibitors demonstrated improved in vitro ADME properties and the ability to induce cytokine production in primary human T-cells.
Organizational Affiliation:
Genentech Inc., 1 DNA Way, South San Francisco, California 94080, United States.